Controlling and optimizing steps in extracting RNA from PAXgene tubes significantly improves yield and quality and can lead to more accurate preclinical and clinical conclusions.
Optimized workflows for PAXgene tubes in clinical studies
Blood samples are a critical component of clinical trials, and they provide essential biological data that can be used to assess the safety and efficacy of new drugs and therapies. They are also used for genetic testing to identify variations that may predict individual responses to treatment.
A key consideration for large, multicenter clinical studies is the ability to store blood samples for later RNA isolation. However, blood plasma is high in ribonuclease (RNase) activity and can rapidly degrade RNA if whole blood samples are isolated, stored, or processed incorrectly, changing the RNA profile within minutes (Baechler et al., 2004). This hinders researchers from detecting and evaluating molecular biomarkers of disease. One of the methods to limit RNA degradation and maintain RNA quality in whole blood is Paxgene tubes (PreAnalytiX). In Paxgene tubes, RNA is stable for three days at room temperature, five days at 4°C, and at least 11 years of storage at -20°C (PreAnalytiX, 2018). In addition, automated sample preparation has become a key factor for many laboratories / GCLP laboratories to guarantee reproducibility of results.
Stability of RNA in frozen and thawed PAXgene tubes
Considering the wide-spread use of freezing Paxgene tubes for RNA isolation, it is essential to assess stability during the transition from frozen to thawed samples. Incorrect thawing may release RNases, with samples differing in sensitivity to RNase activity depending on the cell type and donor. To minimize RNase exposure, it is essential to process samples efficiently, reduce thawing time, and include washing steps to remove any released RNase (Box 1).
Box 1: PAXgene Tube Workflow
Automated extraction of intra-cellular RNA from Paxgene tubes using QIAsymphony PAXgene Blood RNA Kit on the QIAsymphony SP, Qiagen.
- After thawing, RNA isolation begins by a pelleting step of the contents of the PAXgene Blood RNA Tube.
- The pellet is washed, and proteinase K is added to digest proteins.
- Alcohol is added to adjust binding conditions.
- Lysates are centrifuged to remove cell debris.
- Residual DNA is removed by a DNase I digestion, and the remaining contaminants are removed in multiple wash steps before pure RNA is eluted.
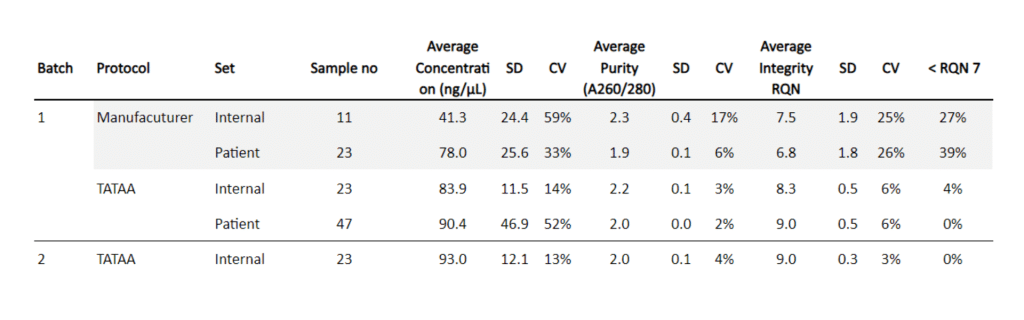
The TATAA optimized PAXgene tube extraction workflow increased average RNA concentration and integrity while reducing variability.
Our optimized protocol for PAXgene tube RNA extraction
TATAA recognizes the importance of stable handling for Paxgene tubes, explicitly focusing on optimizing the workflow for frozen tubes and automated high-throughput processes to meet the demands of clinical sampling.
By optimizing defrosting time and making adjustments to the washing steps, we aimed to increase RNA concentration and integrity to reduce sample variability.
TATAA Biocenter collected 127 human blood samples in total and measured the concentration, purity, and integrity of RNA isolated using either the manufacturer’s standard protocol for automated RNA extraction or a TATAA-optimized protocol. In addition, within the same set of samples, we assessed the robustness of our results by comparing internal samples collected in a controlled manner with patient samples from uncontrolled settings. Internal samples were analyzed in two batches at separate time points and by different scientists.
Internal samples were collected and initially incubated for two hours at room temperature, subsequently stored at +4°C for two days, and transferred to -20°C for two days before storage at -80°C. RNA was eluted in a total volume of 80 µL on a 96well plate, then directly incubated at 65°C for 5 minutes in a thermocycler (T100). Sample concentrations and purities were measured using a spectrophotometer (Lunatic, Unchained labs). Subsequently, samples were analyzed for RNA integrity (RNA quality number: RQN) on a capillary electrophoresis system (Fragment Analyzer, Agilent Technologies Inc).
RQN comparison across PAXgene tube protocols
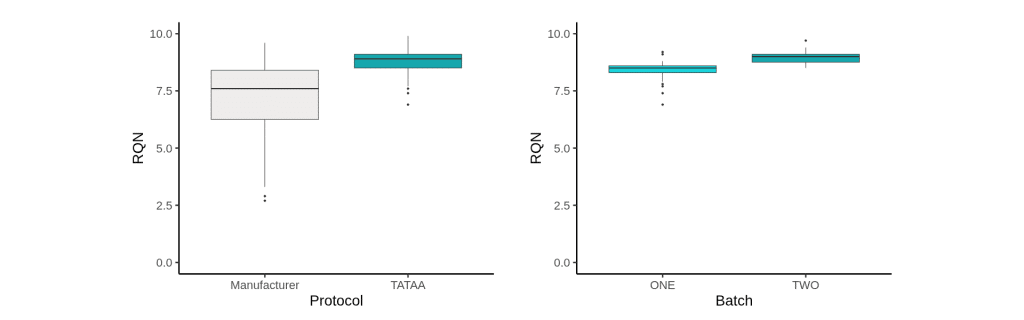
Using PAXgene tubes, the TATAA protocol produced more consistent and higher RNA quality (RQN) than the manufacturer’s protocol, with little difference between batches one and two, demonstrating strong batch-to-batch reproducibility.
TATAA Biocenter expertise in PAXgene tube processing
Starting with PAXgene blood RNA tube samples, TATAA offers a range of tests based on RT-qPCR/RT-dPCR and sequencing of total RNA, mRNA and small RNA.
- Safety monitoring and adverse event detection: We regularly perform blood tests to detect changes in biomarkers that indicate toxicity or potential side effects during treatment.
- Efficacy assessment: Blood samples are analyzed for specific biomarkers that reflect a treatment’s biological effect and efficacy.
- Genetic profiles for personalized medicine: We conduct genetic testing on blood samples to identify variations that may predict individual responses to treatment, enabling personalized therapeutic approaches.
- Biomarker stratification: Blood biomarkers are used to stratify patients into subgroups, improving trial efficiency.